Multi-enzyme Screening Using a High-throughput Genetic Enzyme Screening System
Journal of Visualized Experiments
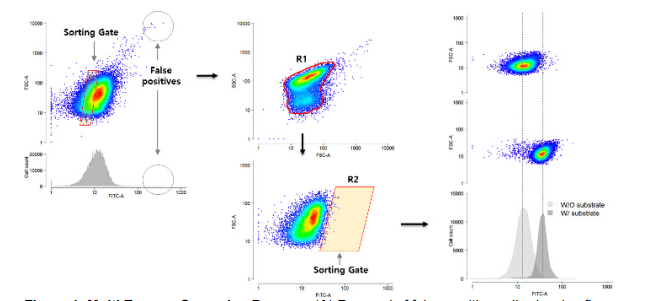 The recent development of a high-throughput single-cell assay technique enables the screening of novel enzymes based on functional activities from a large-scale metagenomic library(1). We previously proposed a genetic enzyme screening system (GESS) that uses dimethylphenol regulator activated by phenol or p-nitrophenol. Since a vast amount of natural enzymatic reactions produce these phenolic compounds from phenol deriving substrates, this single genetic screening system can be theoretically applied to screen over 200 different enzymes in the BRENDA database. Despite the general applicability of GESS, applying the screening process requires a specific procedure to reach the maximum flow cytometry signals. Here, we detail the developed screening process, which includes metagenome preprocessing with GESS and the operation of a flow cytometry sorter. Three different phenolic substrates (p-nitrophenyl acetate, p-nitrophenyl-β-D-cellobioside, and phenyl phosphate) with GESS were used to screen and to identify three different enzymes (lipase, cellulase, and alkaline phosphatase), respectively. The selected metagenomic enzyme activities were confirmed only with the flow cytometry but DNA sequencing and diverse in vitro analysis can be used for further gene identification.
The recent development of a high-throughput single-cell assay technique enables the screening of novel enzymes based on functional activities from a large-scale metagenomic library(1). We previously proposed a genetic enzyme screening system (GESS) that uses dimethylphenol regulator activated by phenol or p-nitrophenol. Since a vast amount of natural enzymatic reactions produce these phenolic compounds from phenol deriving substrates, this single genetic screening system can be theoretically applied to screen over 200 different enzymes in the BRENDA database. Despite the general applicability of GESS, applying the screening process requires a specific procedure to reach the maximum flow cytometry signals. Here, we detail the developed screening process, which includes metagenome preprocessing with GESS and the operation of a flow cytometry sorter. Three different phenolic substrates (p-nitrophenyl acetate, p-nitrophenyl-β-D-cellobioside, and phenyl phosphate) with GESS were used to screen and to identify three different enzymes (lipase, cellulase, and alkaline phosphatase), respectively. The selected metagenomic enzyme activities were confirmed only with the flow cytometry but DNA sequencing and diverse in vitro analysis can be used for further gene identification.
DOI:10.3791/nar/gkac206. IF.(y). Citation . ISSN no.-.