Efficient Transcriptional Gene Repression by Type V-A CRISPR-Cpf1 from Eubacterium eligens
ACS Synthetic Biology
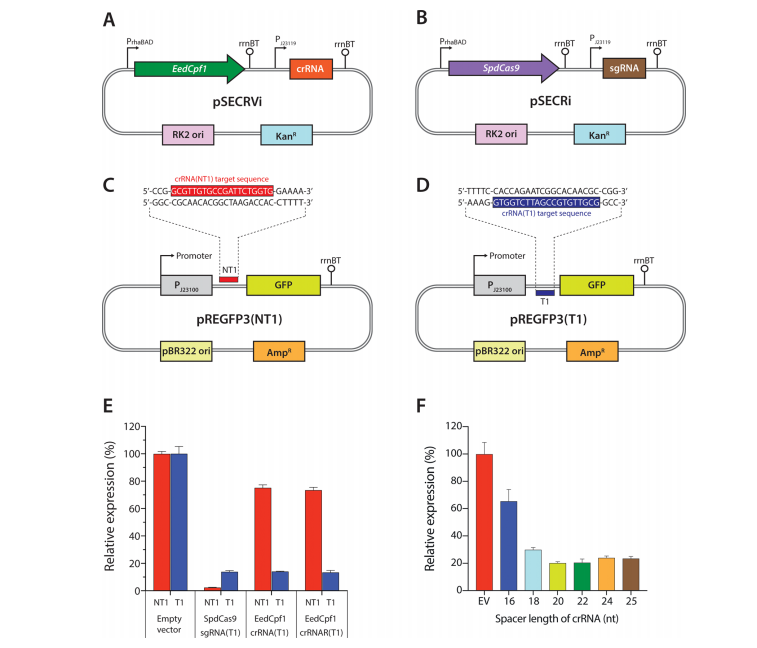 Clustered regularly interspaced short palindromic repeats interference (CRISPRi) is an emerging technology for artificial gene regulation. Type II CRISPR-Cas endonuclease Cas9 is the most widely used protein for gene regulation with CRISPRi. Here, we present type V-A CRISPR-Cas endonuclease Cpf1-based CRISPRi. We constructed an l-rhamnose-inducible CRISPRi system with DNase-deactivated Cpf1 from Eubacterium eligens (EedCpf1) and compared its performance with catalytically deactivated Cas9 from Streptococcus pyogenes (SpdCas9). In contrast to SpdCas9, EedCpf1 showed stronger gene repression when it was targeted to the template strand than when it was targeted to the nontemplate strand of the 5’ untranslated region or coding DNA sequences. EedCpf1 exhibited no strand bias when targeted to the promoter, and preferentially used the 5’-TTTV-3’ (V = A, G, or C) protospacer adjacent motif. Multiplex repression of the EedCpf1-based CRISPRi system was demonstrated using episomal and chromosomal gene targets. Our findings will guide an efficient EedCpf1-mediated CRISPRi genetic control.
Clustered regularly interspaced short palindromic repeats interference (CRISPRi) is an emerging technology for artificial gene regulation. Type II CRISPR-Cas endonuclease Cas9 is the most widely used protein for gene regulation with CRISPRi. Here, we present type V-A CRISPR-Cas endonuclease Cpf1-based CRISPRi. We constructed an l-rhamnose-inducible CRISPRi system with DNase-deactivated Cpf1 from Eubacterium eligens (EedCpf1) and compared its performance with catalytically deactivated Cas9 from Streptococcus pyogenes (SpdCas9). In contrast to SpdCas9, EedCpf1 showed stronger gene repression when it was targeted to the template strand than when it was targeted to the nontemplate strand of the 5’ untranslated region or coding DNA sequences. EedCpf1 exhibited no strand bias when targeted to the promoter, and preferentially used the 5’-TTTV-3’ (V = A, G, or C) protospacer adjacent motif. Multiplex repression of the EedCpf1-based CRISPRi system was demonstrated using episomal and chromosomal gene targets. Our findings will guide an efficient EedCpf1-mediated CRISPRi genetic control.
DOI:10.1021/nar/gkac206. IF.(y). Citation . ISSN no.1273-1282.